1st year in "Integration of results (WP 8)"[]
The value of multiple neuronal-network simulators[]
There are many freely-available, open-source, well-documented tools for simulation of networks of spiking neurons (see this article for a recent review). Each strikes a different balance between efficiency, flexibility and scalability, and the different simulators encompass a range of simulation strategies. Moreover, simulators used in computational neuroscience are complex software packages, and may have unexamined assumptions that may only be apparent in particular circumstances.
Therefore it is desirable that any given model should be simulated using several simulators and the results cross-checked. In general, we believe that the field of computational neuroscience would gain greatly from the ability to easily simulate a model with multiple simulators: reduction of implementation-dependent bugs, better benchmarking, improvement of communication between investigators, etc.
All of this argues for simulator-independent model specification. In order to achieve such a specification we have developed both an extension of the NeuroML declarative framework and a new procedural framework (PyNN), in active consultation with other scientific colleagues.
Building a common knowledge repository: avoiding "not-invented-here" syndrome[]
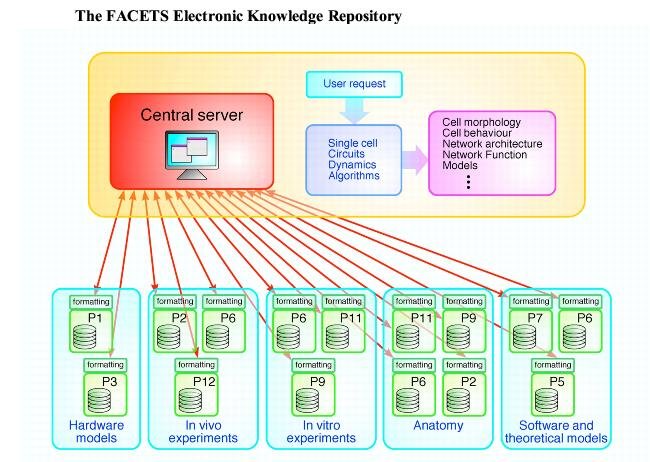
With fifteen partners from biology, hardware and computational science, sharing state-of-the-art, complex and diverse data between colleagues with different levels of expertise in using computer tools, building a common knowledge repository was a challenging issue! Our main principles are not to re-invent what is available elsewhere†, and to focus on making available in standardised formats as much information about the data (meta-data) as possible. This is an ongoing task, with important milestones already reached, and more to come...
†The underlying technology is based on W3C standards and related tools: much functionality comes for free thanks to this choice. The basic platform is built over SRB and data are stored using the HDF5 format. Python is our common scripting language for shared applications, while Java is the basic platform for all utilities.